Decoding Breast cancer: Machine Learning Offers New Prognosis Insights
Table of Contents
- 1. Decoding Breast cancer: Machine Learning Offers New Prognosis Insights
- 2. What are the specific mitochondrial and lysosomal genes that were found to be moast influential in predicting breast cancer prognosis, and how did their expression or mutation status correlate with patient outcomes?
- 3. Decoding Breast cancer: Machine Learning Offers New Prognosis insights
- 4. Interview with Dr. Anya Kapoor,Lead Researcher
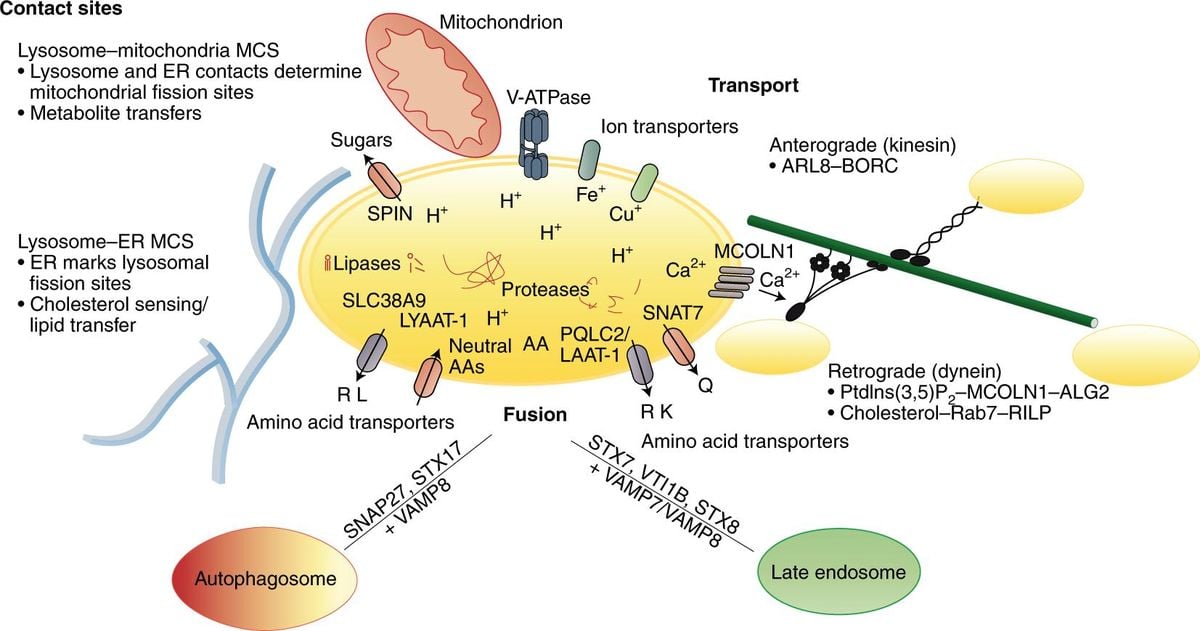
In the ongoing fight against breast cancer, researchers are continually exploring innovative approaches to improve patient outcomes.A groundbreaking study published recently highlights the potential of machine learning to revolutionize breast cancer prognosis. By delving into the intricate interplay of mitochondrial and lysosomal genes, scientists have developed powerful algorithms capable of predicting patient risks and guiding personalized treatment strategies.
Traditionally, understanding the role of mitochondrial and lysosomal functions in breast cancer has been clouded by ambiguity. This study seeks to shed light on this intricate relationship, analyzing vast datasets from multiple sources. Through sophisticated techniques like differential expression analysis and copy number variation assessments, researchers identified key prognostic markers associated with breast cancer.
A significant finding of the study underscores the crucial role of B-cell immune infiltration in patient survival. Reduced B-cell infiltration was strongly linked to poorer outcomes, opening up exciting avenues for potential therapeutic targets. As the authors stated, “This study shows the machine learning model demonstrated strong associations with patient outcomes,” highlighting the transformative power of this integrated approach.
The study’s rigorous methodology involved analyzing data from 4,897 breast cancer patients across multiple datasets, rigorously validating the model’s predictive accuracy. It emphasizes the importance of comprehensively evaluating mitochondrial and lysosomal gene activities to fully grasp their influence on tumor biology.This resonates with the complex nature of breast cancer, frequently enough characterized by genetic variations and resistance mechanisms.
Previous research has established a link between elevated photon metabolism and mitochondrial dysfunction, a phenomenon frequently associated with treatment resistance. researchers employed advanced statistical methods like univariate Cox regression and machine learning algorithms such as CoxBoost and survival-SVM to stratify patients more effectively than conventional methods. This paves the way for identifying high-risk patient cohorts who require immediate and tailored therapeutic interventions.
The implementation of sophisticated machine learning models, like the one developed in this study, signifies a crucial leap forward in precision medicine within oncology. “Enhancing B cell infiltration and mitochondrial lysosome activity emerges as personalized interventions for high-risk patients,” states the study, emphasizing the profound impact on clinical decision-making.
This study serves as a powerful testament to the growing potential of machine learning in genomics. It not only provides valuable research insights but also offers tangible clinical applications for improving breast cancer management. The key takeaway is the potential for predictive models to subtly yet significantly shift the landscape of cancer care. by recognizing mutations and cellular behaviors that influence treatment response,these models pave the way for more precise and effective therapies.
Moving forward, further research and validation through clinical trials are crucial to solidify the applicability of these models across diverse patient populations. This study lays a solid foundation for future endeavors aimed at developing robust and evidence-based tools for breast cancer prognosis, ultimately translating the knowledge gained from mitochondrial and lysosomal interactions into improved patient outcomes.
What are the specific mitochondrial and lysosomal genes that were found to be moast influential in predicting breast cancer prognosis, and how did their expression or mutation status correlate with patient outcomes?
Decoding Breast cancer: Machine Learning Offers New Prognosis insights
In the ongoing fight against breast cancer, researchers are continually exploring innovative approaches to improve patient outcomes.A groundbreaking study published recently highlights the potential of machine learning to revolutionize breast cancer prognosis. By delving into the intricate interplay of mitochondrial and lysosomal genes, scientists have developed powerful algorithms capable of predicting patient risks and guiding personalized treatment strategies.
Interview with Dr. Anya Kapoor,Lead Researcher
Archyde: Dr. Kapoor, your research exploring the link between mitochondrial and lysosomal genes and breast cancer prognosis has generated important buzz. can you tell us about the inspiration behind this study and its primary objectives?
Dr. Kapoor: Thank you for having me. The field of oncology has long recognized the crucial role of mitochondria in tumor biology. However, the intricate relationship between mitochondrial function, lysosomal activity, and breast cancer progression remained poorly understood. Our primary objective was to unravel this complex interplay and determine if we could harness this knowledge to improve patient prognosis and treatment strategies.
Archyde: What were the key methodologies used in this study, and what were some of the most striking findings?
Dr. Kapoor: We employed a multi-pronged approach. We analyzed vast datasets from over 4,897 breast cancer patients, utilizing refined techniques like differential expression analysis and copy number variation assessments to identify key prognostic markers associated with patient survival. Notably, we found that reduced B-cell immune infiltration was strongly linked to poorer outcomes, highlighting the potential of targeting B-cells as a therapeutic strategy.
Archyde: Machine learning played a central role in your research. How did these algorithms contribute to the study’s insights, and what do these findings suggest for personalized medicine in breast cancer?
Dr.Kapoor: our machine learning models were trained on extensive genomic and clinical data, enabling them to identify complex patterns and correlations that might have gone unnoticed with traditional methods. This allowed us to develop a predictive model that stratifies patients based on their risk profiles, possibly enabling more targeted and personalized treatments. As an example, patients identified as high-risk could receive intensified therapy or early interventions, while those at lower risk might benefit from less aggressive approaches.
Archyde: What are the next steps for your research, and what could these findings mean for the future of breast cancer treatment?
Dr. Kapoor: We are currently conducting further validation studies and exploring the potential of these models in clinical settings. Our long-term goal is to develop clinically actionable tools that integrate mitochondrial and lysosomal gene activity as part of routine breast cancer diagnostics and treatment planning. This could lead to more precise therapies and ultimately improve patient outcomes.
Archyde: This research offers a glimmer of hope for countless individuals battling breast cancer.What message would you like to share with patients reading this?
Dr. Kapoor: We are making significant strides in understanding the complexities of breast cancer. While there is still much work to be done, these findings demonstrate the groundbreaking potential of machine learning and genomic research in revolutionizing personalized cancer care. Stay informed about the latest advancements, engage actively with your healthcare providers, and never lose hope.